Abstract
Introduction
Flow cytometry (FC) is a reliable tool for minimal residual disease (MRD) detection in acute leukemia. However, manual interpretation inevitably suffers from individual idiosyncrasy even by experienced physicians, which could affect diagnostic reproducibility and objectivity. With recently advanced artificial intelligence (AI) technology, we aim to develop an automated FC data interpretation algorithm to support physicians in objectively detecting MRD for acute myeloid leukemia (AML) and myelodysplastic syndrome (MDS).
Methods
From 2009 to 2017, 4350 AML or MDS FC data samples for MRD detection at National Taiwan University Hospital were enrolled, from two different machines (3090 from Calibur and 1260 from CantoII-Senior). A formula of inter-machine value calibration was provided by the manufacturer. A 12-tube test was performed for each sample: 100000 cells measured in 6 fluorescent channels (FSC, SSC, FITC, PE, PerCP, APC) per tube.
The whole dataset was randomly divided into Training set 1, Training set 2 and Validation set, taking 64%, 16% and 20% samples respectively. The Training set 1 was for developing AI algorithm, and the Training set 2 for parameter tuning. Final concordance was estimated with the blinded Validation set. Algorithms for three paired comparisons (AML vs. normal, MDS vs. normal, and abnormal (AML+MDS) vs. normal) were developed independently, according to previous manual analysis.
For algorithm development, the recorded numerical values of the 6 aforementioned fluorescent channels of each tube were used as raw feature attributes. The probabilistic distributions of these attributes was modeled as multivariate Gaussian mixture model by a sub-dictionary learning approach. A probabilistic derivation was then exploited to compute per-sample L2-normalized vectorized representations. Lastly, representations of each tube were then concatenated to be the final feature input to the supervised machine learning classifier, i.e., a support vector machine with linear kernel. ANOVA-based feature selection was also conducted.
For clinical validation of AI-assisted FC analysis, clinical characters and outcomes from 120 AML patients with available FC data after standard induction were analyzed, including their overall survival (OS) and progression-free survival (PFS).
Results
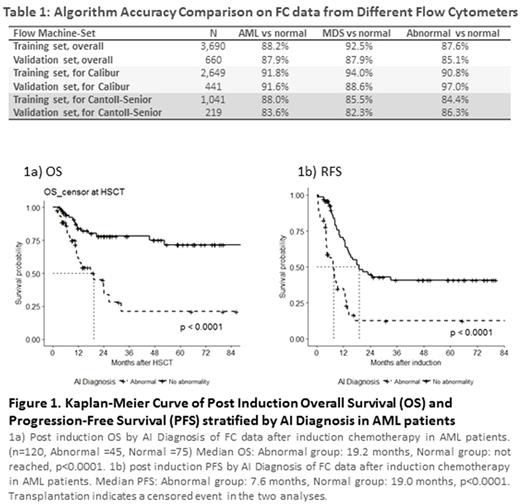
As in Table 1, AI algorithm achieved promising concordance rates of 88.2%, 92.5%, and 87.6% in pair-wise recognition tasks, i.e., AML vs normal, MDS vs normal, and abnormal vs normal, respectively. Similar results were noted for the Validation set, with the rates of 87.9%, 87.9%, and 85.1%. The areas under receiver operating characteristic curve (AUCs) were 0.903, 0.784 and 0.886, respectively. Concordance was reduced when data from different machines were aggregated even with conversion formula, indicating that algorithm may need adjustment for different machines.
It is particularly noteworthy that only 7 seconds were required for AI to interpret one sample, i.e., at least 130 folds faster than current manual work.
Moreover, even with only 1 tube data, high concordance was maintained. When comparing abnormal vs. normal in Calibur data, algorithm from all 12 tube data yielded an AUC of 0.935, which remained unchanged as tube number decreased. In fact, the AUC with only 1 tube data (CD13, CD16, CD45, FSC and SSC) was 0.915. This was also noted in CantoII-Senior data (AUC 0.832 and 0.790 for 12 and 1 tube), and across all other comparisons. The findings implicate that AI may reduce laboratory works currently required for high-fidelity manual analysis.
For the 120 AML cases with available post-induction FC data, 45 were classified as abnormal (i.e., positive MRD) and 75 as normal by AI algorithm. As in Figure 1, patients with abnormal FC by AI had a significantly worse PFS (median 7.6 vs. 19.0 months, p<0.0001), and OS (median 19.2 months vs. not reached, p<0.0001) than those with normal FC. Multivariate analysis with Cox hazard proportional model confirmed the outcome predicting significance for AI interpretation.
Conclusions
We demonstrated reliable accuracies and efficient prognostic stratification for AI-assisted MRD detection in AML/MDS, with high-speed interpretation and labor saving. Further integration with other laboratory or genomic data into AI algorithm may facilitate more precise outcome prediction and risk-adapted intervention for these patients.
Ko: Celgene International Sàrl: Research Funding. Li: Celgene International Sàrl: Research Funding. Hou: Celgene International Sàrl: Research Funding. Tien: Celgene International Sàrl: Research Funding. Tang: Celgene International Sàrl: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal